Generative Models for Synthetic Biosignal Data
Leveraging GANs and Diffusion Models to synthesize high-fidelity biomedical time-series data for data augmentation.
Biomedical signal processing is a cornerstone of modern healthcare and assistive technologies. However, machine learning applications in this domain are often hampered by significant data challenges. Datasets for signals like electrocardiograms (ECG) or electroencephalograms (EEG) are typically small, expensive to collect and annotate, and subject to strict privacy constraints. Furthermore, these datasets are frequently imbalanced, with critical but rare event data being underrepresented, which biases model training.
This project addresses these challenges by developing and evaluating deep generative models capable of creating high-fidelity, realistic synthetic biosignals. By leveraging the power of Generative Adversarial Networks (GANs) and Denoising Diffusion Probabilistic Models (DDPMs), our work provides a robust framework for data augmentation, class imbalance correction, and data denoising, ultimately enhancing the performance and reliability of downstream machine learning models.
From GANs to Transformers
Our initial approach focused on Generative Adversarial Networks. We progressed from standard RNN-based models to more advanced, transformer-based architectures that are better suited for capturing long-range dependencies in time-series data.
TTS-GAN (Li et al., 2022) introduced a pure transformer-based GAN capable of generating realistic single-class time-series data. To address multi-class generation and the issue of data scarcity in minority classes, we developed BioSGAN (Li et al., 2023), a conditional, label-guided model. BioSGAN can be trained on an entire multi-class dataset, leveraging transfer learning between classes to generate high-quality, class-specific signals, even for classes with very few real examples.
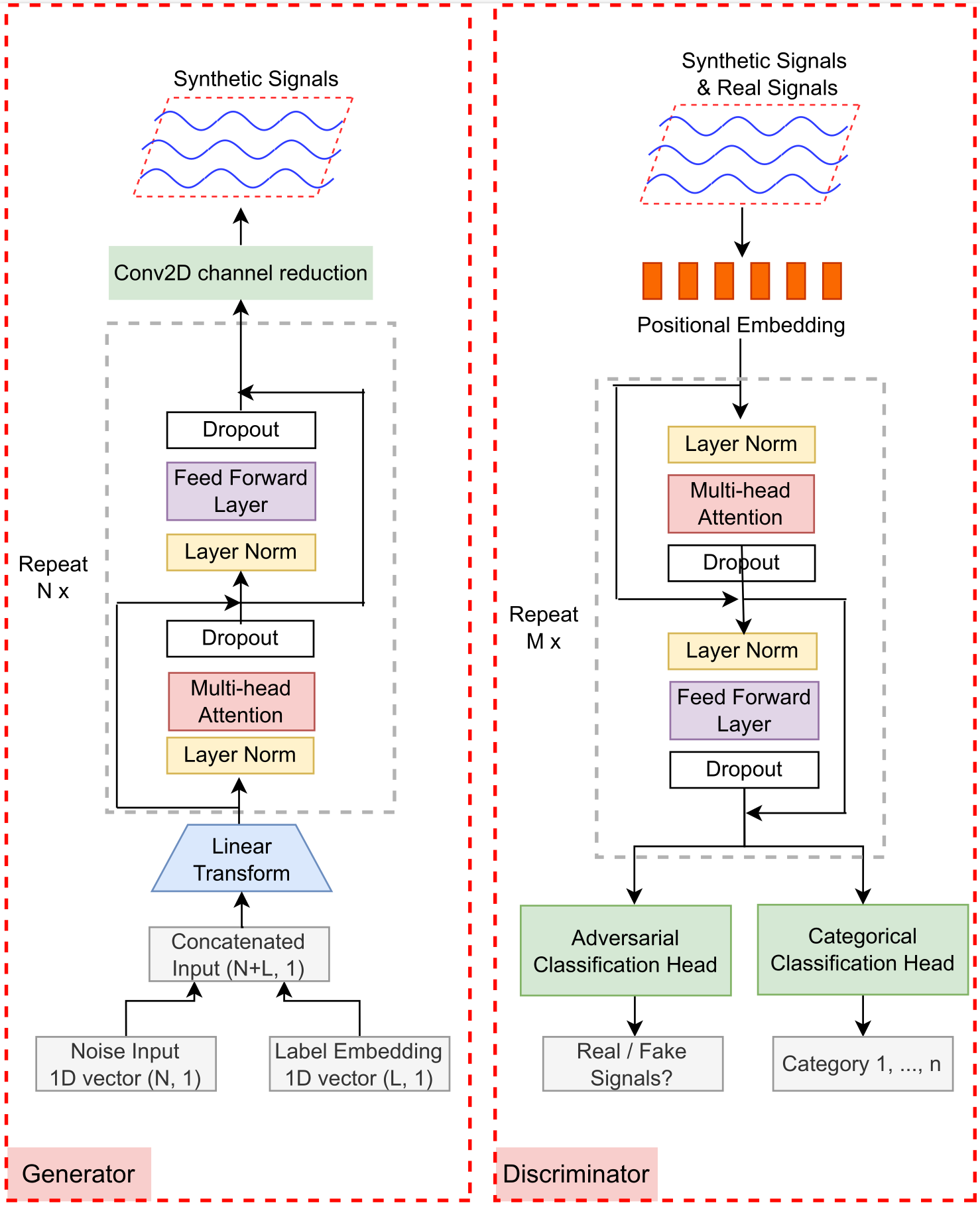
Advancing with Diffusion Models
While GANs are powerful, Diffusion Models have recently emerged as a state-of-the-art alternative, often producing more diverse and high-fidelity samples. Our research has explored their application to biosignal synthesis with a focus on versatility and robustness.
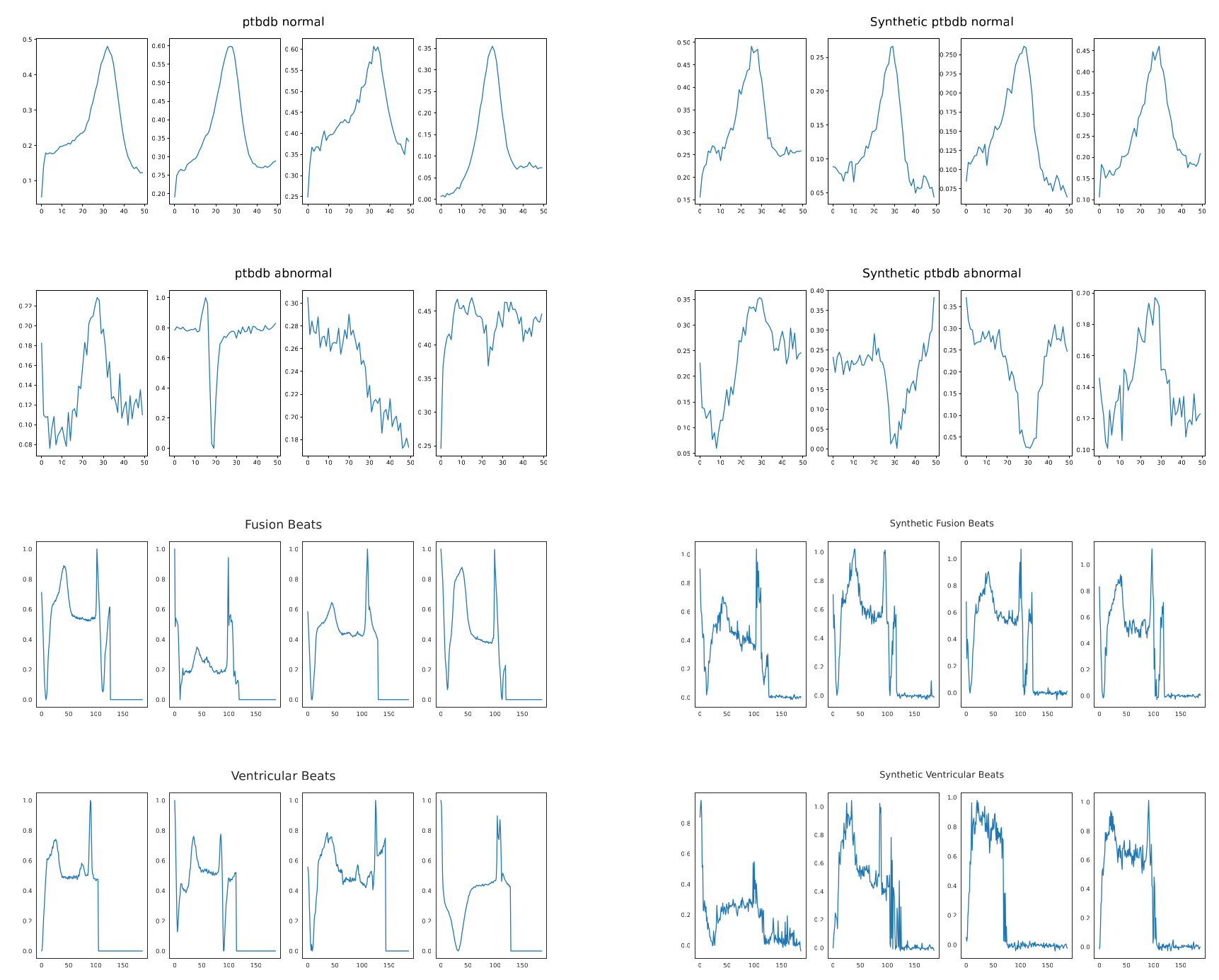
BioDiffusion (Li et al., 2024) is a versatile framework for unconditional, label-conditional, and signal-conditional generation. This single model can not only generate new data from scratch but can also perform tasks like denoising, signal imputation (filling in missing values), and super-resolution, making it a comprehensive tool for signal enhancement.
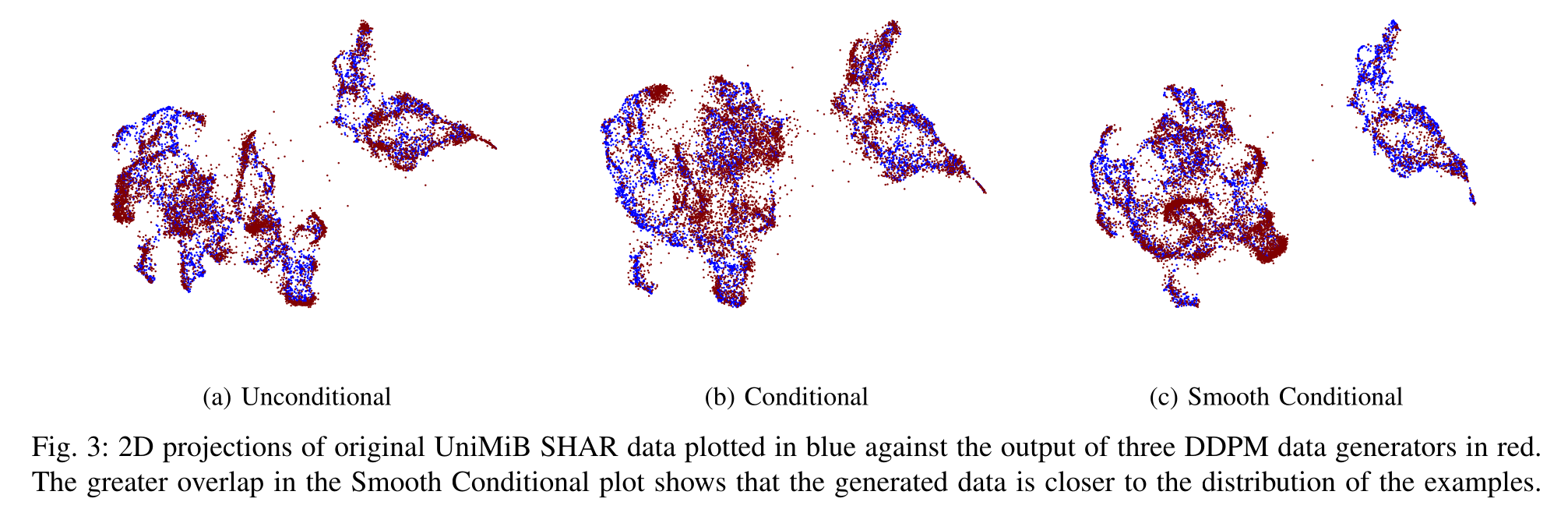
Furthermore, we tackled the real-world problem of noisy labels in training data. Our work in (Atkinson et al., 2023) introduces a novel adaptation of Denoising Diffusion Probabilistic Models (DDPMs) that incorporates label smoothing. This technique prevents the model from becoming overconfident in mislabeled data, significantly improving the quality and reliability of the generated signals when trained on imperfect datasets.
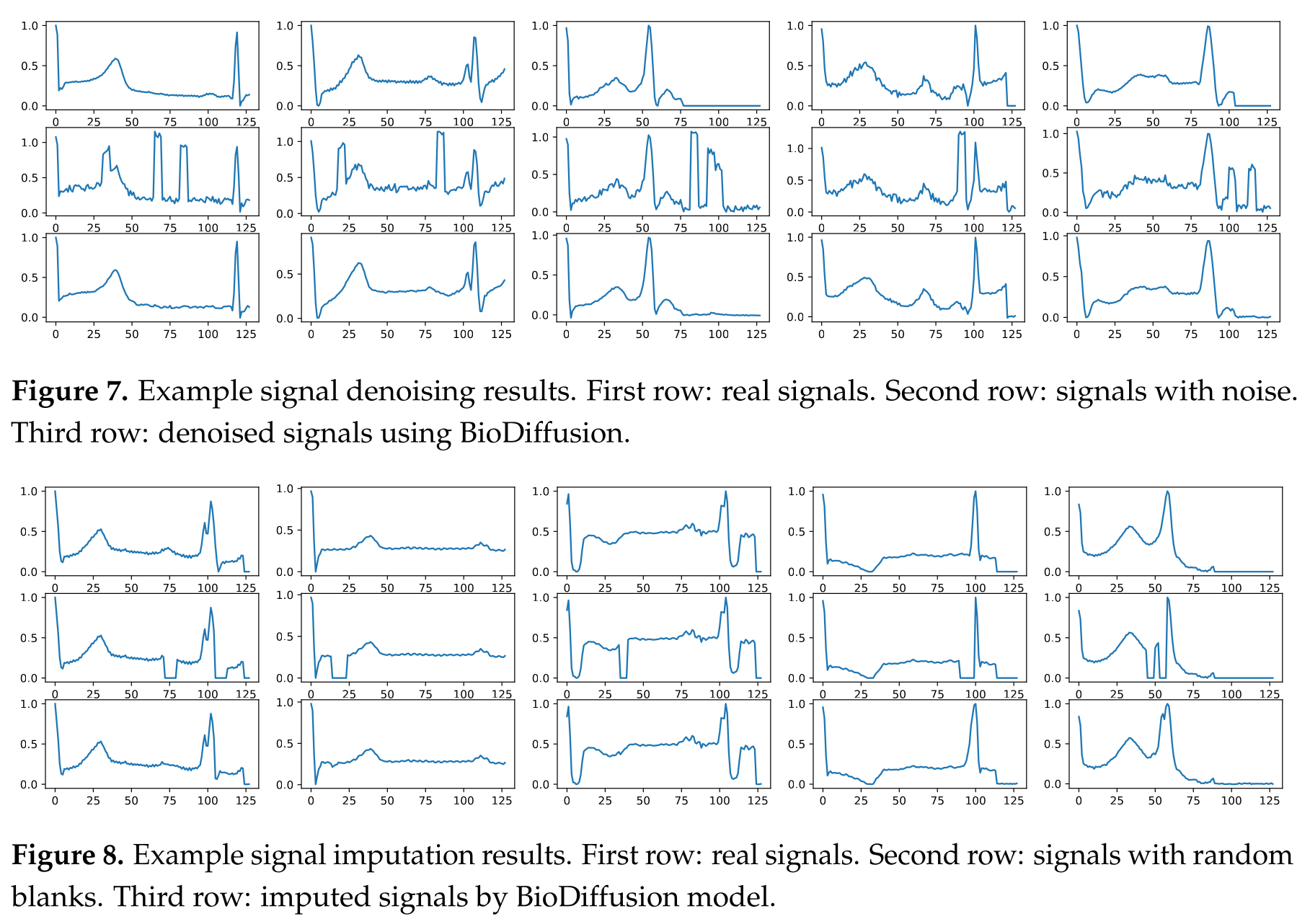
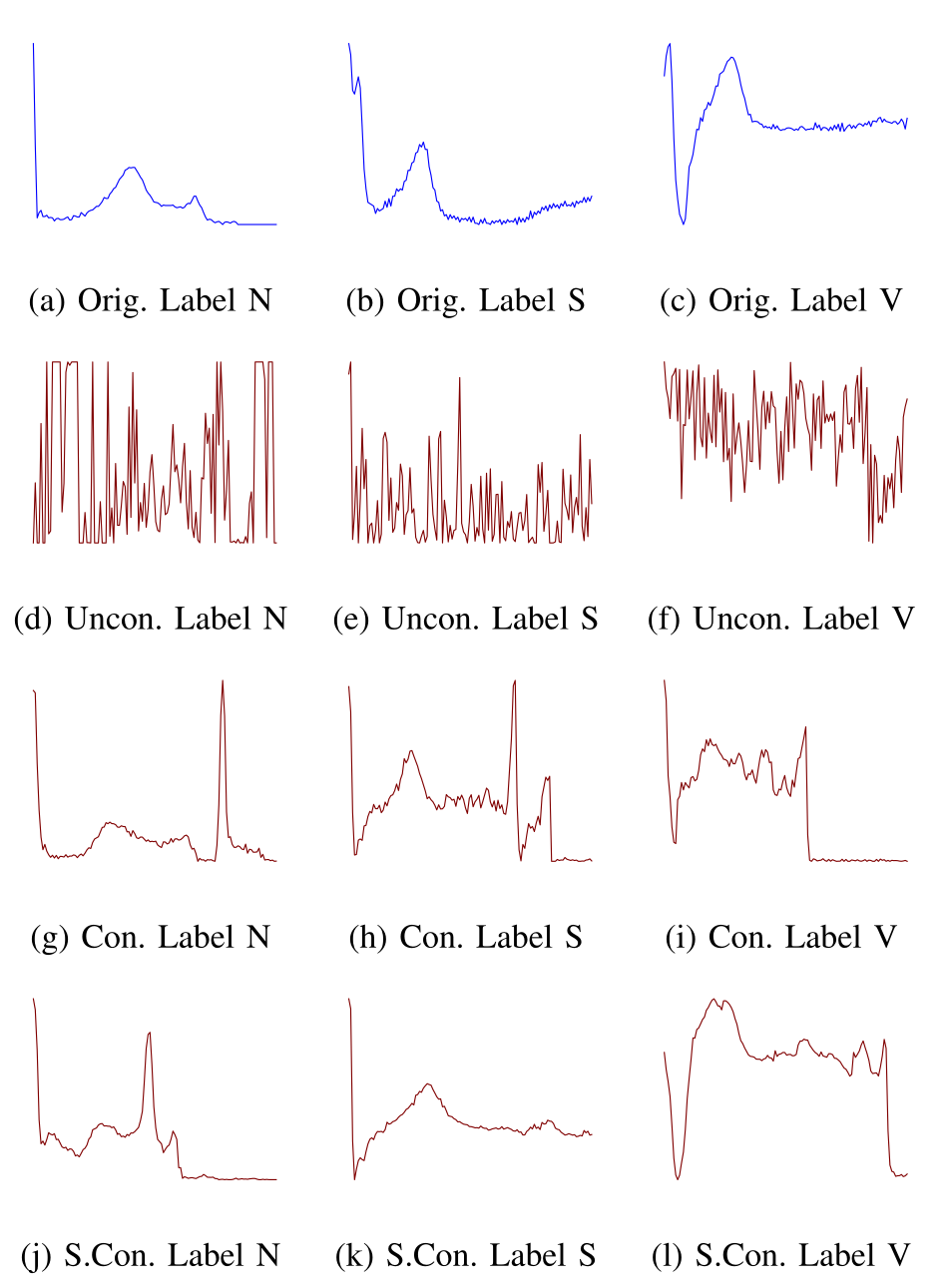
Impact on Downstream Tasks
The ultimate goal of generating synthetic data is to improve the performance of other machine learning models. We have demonstrated that augmenting imbalanced datasets with our synthetically generated signals significantly improves the performance of classifiers. For instance, in classifying arrhythmia from ECG signals, adding synthetic data for rare classes dramatically increased the F1-score and recall for those classes, leading to a more robust and clinically useful diagnostic model.
The quality of the generated data is validated through both quantitative metrics and qualitative visualizations, such as the t-SNE plot below, which shows a strong overlap between the distributions of real and synthetic data.